Introduction to C-N Hydrolases
 The term "carbogen-nitrogen hydrolase" (C-N hydrolase) was introduced by Peer Bork and Eugene Koonin in 1994
to describe a new protein superfamily (Bork and Koonin,
1994). They compared 15 sequences which were classified in 6 different groups and while some members of this family were only very distantly
related, all of them had in common that they were catalyzing the hydrolytic cleavage of C-N bonds. It should be mentioned that although all C-N
hydrolases are cleaving some type of C-N bonds, not every C-N-bond-cleaving enzyme is a member of this superfamily. Proteases, for example,
are also cleaving C-N bonds (within proteins), but none of the proteases currently known is phylogenetically related to the C-N-hydrolase family.
The term "carbogen-nitrogen hydrolase" (C-N hydrolase) was introduced by Peer Bork and Eugene Koonin in 1994
to describe a new protein superfamily (Bork and Koonin,
1994). They compared 15 sequences which were classified in 6 different groups and while some members of this family were only very distantly
related, all of them had in common that they were catalyzing the hydrolytic cleavage of C-N bonds. It should be mentioned that although all C-N
hydrolases are cleaving some type of C-N bonds, not every C-N-bond-cleaving enzyme is a member of this superfamily. Proteases, for example,
are also cleaving C-N bonds (within proteins), but none of the proteases currently known is phylogenetically related to the C-N-hydrolase family.
In more recent literature, the term "nitrilase superfamily" is often used instead of "C-N hydrolase" (Pace and Brenner, 2001;
Brenner, 2002). This is because many members of this superfamily
were assigned to be "nitrilases" or "nitrilase-like" just due to their sequence similarity to known nitrilase sequences. Today it is clear that many
of these proteins are not nitrilases, but often their exact enzymatical function is still unknown.
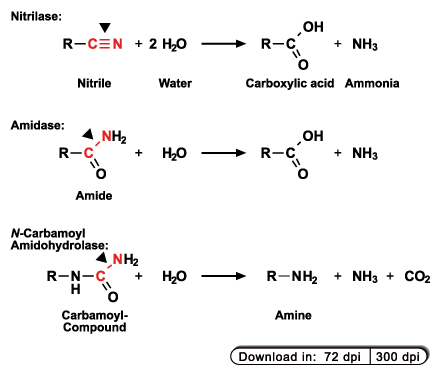
C-N hydrolases are distributed throughout the tree of life; they can be found in bacteria as well as in men and were classified into 13 different
functional groups. As an example, three different types of C-N hydrolases, namely nitrilases, aliphatic amidases, and carbamoyl
amidohydrolases and the reactions typically catalyzed by them are shown in the figure on the right. As can also be seen, the release of ammonia
is usually associated with C-N hydrolase reactions.
In our laboratory we are working on nitrilases of plants and on N-carbamoylputrescine amidohydrolase, an enzyme involved in the biosynthesis
of polyamines.
C-N Hydrolases of Arabidopsis thaliana and Oryza sativa
The fully sequenced genomes of the two plants Arabidopsis thaliana and Oryza sativa each contain six different classes of C-N-hydrolases:
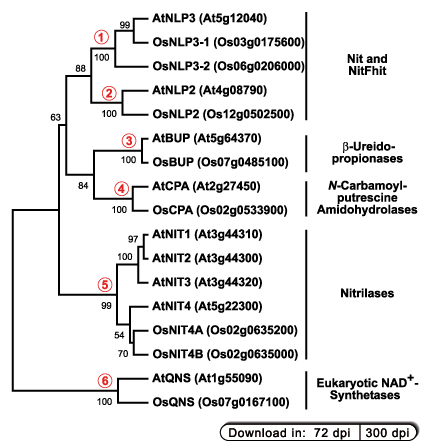
1. + 2. Nitrilase-like proteins (NLP) of unknown functions. According to the classification of Pace and Brenner (2001)
they belong to the "Nit and NitFhit" branch.
3. β-Ureidopropionases (syn. β-alanine synthases) catalyze the formation of β-alanine or
β-aminoisobutyrate from β-ureidopropionate (a catabolite of uracil) or β-ureidoaminobutyrate (a catabolite of thymine), respectively. β-Alanine
is a precursor of pantothenate (vitamin B5) which is a constituent of coenzyme A.
The cDNA for the β-ureidopropionase of Arabidopsis thaliana has been cloned and the recombinant enzyme was biochemically characterized (Walsh
et al., 2001).
4. N-Carbamoylputrescine amidohydrolase is involved in the biosynthesis of the parent polyamine putrescine. The two
CPA enzymes of Arabidopsis thaliana and Lycopersicon esculentum have been expressed in
Escherichia coli and were subsequently characterized (Piotrowski
et al., 2003).
5. Nitrilases catalyze the hydrolysis of nitriles into their carboxylic acids (see also figure above). The NIT4 homologs metabolize β-cyanoalanine,
an intermediate product in the cyanide detoxification pathway of higher plants, to aspartic acid and asparagine (Piotrowski
et al., 2001; Piotrowski and Volmer, 2006).
Homologs of NIT1, NIT2 and NIT3 are restricted to the Brassicaceae and may be involved in the catabolism of glucosinolates (Vorwerk et al., 2001).
6. Glutamine-dependent eukaryotic NAD+ synthetase catalyze the final step in the biosynthesis of the coenzyme NAD+. In these enzymes
the C-N-hydrolase function is located in the N-terminal part and serves to release NH3 from glutamine, which is subsequently added to the NAD+
precursor NAAD+.