The EU-FP7 project MATCHIT (Matrix for Chemical IT) explored the construction of a second level production chemistry, which could serve in future as a matrix for future self-reconstructing systems, involving the autonomous chemical manipulation of packets of chemicals, using information tagged chemical containers (chemtainers). EU page for MATCHIT.
"We introduce an integrated programmable information- and production chemistry by having DNA addressable chemical containers (chemtainers) interfacing traditional electronic computers via microelectromechanical systems (MEMS) with regulatory feedback loops. DNA tags anchored in the chemtainers make them addressable with respect to each other through complementary DNA interaction as well as addressable within a MEMS microfluidics matrix through DNA tags anchored in the micro fluidics channels.”
BioMIP was one of 6 partners from 3 institution involved in this pioneering initiative in the Future Emerging Technologies program. The role of BioMIP was primarily to develop electronic microfluidic systems to interact with and program the basic functions of the chemical matrix. BioMIP focussed on three chemtainer aspects:
- the addressing of chemtainers to specific locations within microfluidic structures for electronic processing
- iterative processing of chemtainers requires the selective extraction and packaging of chemtainer reaction products in fresh chemtainers
- on-going iterative processing requires informational reaction products to be amplified to chemtainer quantities.
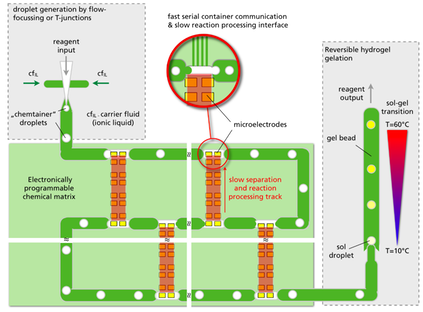
Fig. 1 One possible integration scheme for a Chem-IT matrix involving a fast serial communication track (green) and slow (orange and in this case) gel based reaction processing tracks. Iterative processing of material is made possible via consecutive delivery of container contents to vertical gel segments (orange) that allow transfer to upstream containers.
Aqueous droplets and vesicles were used as chemtainers by BioMIP. Vesicles were labelled as in WP2 (Hanczyk, SDU) using streptavidin-biotin linked DNA and duplex (and in our work pH switchable triplex) DNA structures, and addressed to specific electrodes on microfluidic chips which had been electronically programmed to attach specific matching DNA (publication to appear 2015). The molecular labelling of droplets is more involved, but has the potential to allow the extension of a wide range of droplet processing technologies to the MATCHIT matrix. The approach explored by BioMIP in MATCHIT was to use DNA tags within a droplet to influence the transport properties of a droplet over specific locations (DNA-labelled micro electrodes) on a microfluidic chip. First, droplet braking chips were designed and tested that allow droplets to pause autonomously at specific locations, and be displaced by later droplets. The braking could be tuned to depend sensitively on the hydrophobiicity of the braking location. Next, hydrophilic DNA beads were designed, that could only hybridise to DNA on the surface of the chip when specific DNA adaptor strands were added to the aqueous droplets containing the beads. The braking of such DNA beads at specific DNA electrodes was observed, and the this could be used in future to address aqueous droplets via specific internal labels to specific locations. This is a novel concept in chemtainer processing.
In collaboration with RUB-BioOrg (V. Patzke, G. von Kiedrowski) a two droplet stream amplification scheme for DNA was developed, with only pyrimidine ligation building blocks available during the project.
Further work included the development of a feedback wave electrophoresis technique for adaptive integrated separation and purification of products for iterative chemtainer processing.