Decoy Database Builder
Introduction to the use of the Decoy Database Builder
The false positive rate of protein searches can be estimated by searching decoy databases containing entries with 'right' (original / source) and 'false' (decoy) protein sequences. The false positive rate of a protein identification is defined as the quotient of the number of erroneous identified proteins (from the decoy part) and the sum of the number of all correctly identified proteins (from the source part) and the number of all erroneous identified proteins.
Decoy databases can be created from Fasta formatted protein database files in different ways:
- reverse: the protein sequence of the source database entry is simply reversed.
- shuffle: all amino acids from the source sequence are put in a random sequence.
- random: the mass of the source protein is calculated and a random protein of the same mass (considering a given mass tolerance) is calculated with the same probabilities of amino acid occurrence as the source protein.
Graphical user interface
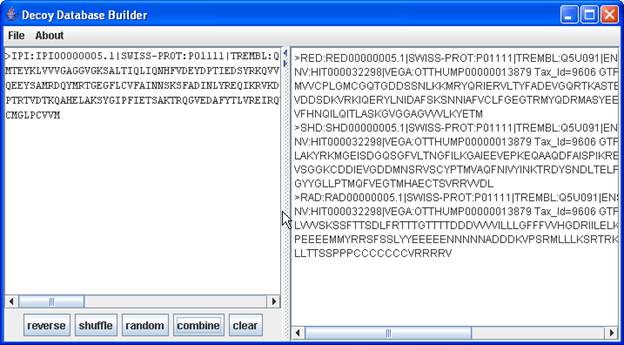
- Source textfield:
- on the left side of the frame
- to enter protein sequences for which decoy sequences should be build
- Buttons beneath the source textfield:
- reverse : for each source sequence a reversed sequence is built from the source textfield and displayed in the Decoy Database textfield on the right.
- shuffle : same as reverse Fasta..., but a shuffled database is built.
- random : same as reverse Fasta..., but a random database is built.
- combine : for each source sequence all decoy sequences are built
- clear : removes all entries from both textfields.
- Decoy Database textfield:
- on the right side of the frame
- here the resulting sequences are displayed
- File menu:
- open Fasta... : a Fasta file can be opened and is displayed in the source textfield
- reverse Fasta... : a reversed version of the database is saved in the same directory as the source database. To distinguish the databases '_reverse' is appended to the original filename before the ‘.fasta’ suffix.
- shuffle Fasta... : same as reverse Fasta..., but a shuffled database is built. ‘_shuffle’ is appended to the original filename.
- random Fasta... : same as reverse Fasta..., but a random database is built. ‘_random’ is appended to the original filename.
- reverse & append Fasta… : both the reversed and the source protein sequences are saved in a new Fasta file. The resulting file is saved with the same name as the original file with the suffix ‘_reverse _decoy.fasta’
- shuffle & append Fasta… : both the shuffled and the source protein sequences are saved in a new Fasta file with the suffix ‘_shuffle _decoy.fasta’.
- random & append Fasta… : both the random and the source protein sequences are saved in a new Fasta file with the suffix ‘_random _decoy.fasta’.
- combine & append Fasta… : all decoy versions and the source protein sequences are saved in a new Fasta file with the suffix ‘_combine _decoy.fasta’.
- Exit : This menu item can be used to close the window in addition to the button in the upper right corner.
Command line interface
If you want to run the software in a larger bioinformatics infrastructure then the DecoyDBB can now be called from within batch scripts. The syntax for generating decoy databases is as follows:
java -jar DecoyDBB filename decoytype append masstolerance
- java: the path to the java interpreter (java.exe) installed on your computer system
- -jar: command line option of the java interpreter to specify that the following java program resides in a java archive (JAR)
- DecoyDBB: the name of the Decoy Database Builder JAR file (without the JAR extension). Make sure that the DecoyDBB.jar file is in the current directory.
- filename: the complete path to the protein database file for which a decoy DB should be generated.
- decoytype: a number to specify the type of the decoy database:
- 0: reverse
- 1: shuffle
- 2: random
- 3: combine (reverse+shuffle+random)
- append: should a hybrid decoy database be generated?
- true: the source and the decoy proteins are contained in the decoy database
- false: only the decoy proteins are contained
- masstolerance: for random (and combined) databases a mass tolerance has to be specified
Formatting rules
- Sequences may use small or upper case letters. The resulting sequences are all upper case.
- Before building the decoy sequences all invalid characters are filtered to ensure a robust behavior of the algorithms.
- In some Fasta formatted protein databases more than the usual 21 amino acids (including selenocysteine) appear. For building a reverse or shuffle decoy database the additional characters B, U, X and Z are not filtered and also appear in the resulting decoy database in reversed or shuffled order. The filtering is still necessary for building random decoy databases, because the mass of those amino acid placeholders can not be correctly calculated.
- To distinguish the decoy from the source proteins the database identifier in the protein accession has to be changed.
- IPI decoy accession identifiers:
- shuffled decoy proteins accessions begin with 'SHD'
- reversed decoy proteins accessions begin with 'RED'
- random decoy proteins accessions begin with 'RAD'
- the source proteins accessions stay the same, i.e. begin with 'IPI'
- NCBI decoy accession identifiers:
- shuffled decoy proteins accessions begin with 'sh'
- reversed decoy proteins accessions begin with 're'
- random decoy proteins accessions begin with 'ra'
- the source proteins accessions stay the same, i.e. begin with 'gi'
- UniprotKB / SwissProt decoy accession identifiers:
- shuffled decoy proteins accessions begin with 'sh'
- reversed decoy proteins accessions begin with 're'
- random decoy proteins accessions begin with 'ra'
- the source proteins accessions stay the same, i.e. begin with a capital letter and a digit.
Examples:
- NCBI example shuffled:
- >gi|385869|gb|AAB26507.1| type XI collagen alpha 1 chain [cattle, vitreous humor, Peptide Partial, 26 aa, segment 7 of 7]
GLPGTQGSPGAGDGGIPGPAGPIGPP
- >sh|385869|gb|AAB26507.1| type XI collagen alpha 1 chain [cattle, vitreous humor, Peptide Partial, 26 aa, segment 7 of 7]
PQGDPPPAGPGLSGAGIITGGGPGPG
- IPI example random:
- >IPI:IPI00000001.1|SWISS-PROT:O95793-1 Tax_Id=9606 Splice Isoform Long of Double-stranded RNA-binding protein Staufen homolog 1
MSQVQVQVQNPSAALSGSQILNKNQSLLSQPLMSIPSTTSSLPSENAGRPIQNSALPSAS...
- >RAD:IPI00000001.1|SWISS-PROT:O95793-1 Tax_Id=9606 Splice Isoform Long of Double-stranded RNA-binding protein Staufen homolog 1
QSQLNSQLMAQPGAQQSKNPKLSNGLQNVLPRQSPPSNVQGNPPRSSQVQSNRPWW...
- UniProtKB / SwissProt example reverse:
- >Q43495|108_SOLLC Protein 108 precursor - Solanum lycopersicum (Tomato) (Lycopersicon esculentum)
MASVKSSSSSSSSSFISLLLLILLVIVLQSQVIECQPQQSCTASLTGLNVCAPFLVPGSPTASTECCNAVQSINHDCMCN
TMRIAAQIPAQCNLPPLSCSAN
- >re3495|108_SOLLC Protein 108 precursor - Solanum lycopersicum (Tomato) (Lycopersicon esculentum)
NASCSLPPLNCQAPIQAAIRMTNCMCDHNISQVANCCETSATPSGPVLFPACVNLGTLSATCSQQPQCEIVQSQLVIVLL
ILLLLSIFSSSSSSSSSKVSAM
in case of any question do not hesitate to contact kai.reidegeld@ruhr-universitaet-bochum.de