Research interests
Yeast SNAREs
Bakerís yeast (Saccharomyces cerevisiae) is used as a model organism because basic cellular functions are very similar in yeast and humans. For technical reasons it is easier and faster to study yeast as a combination of genetic and biochemical approaches can be used. Results obtained in yeast can be applied to mammals in subsequent projects.
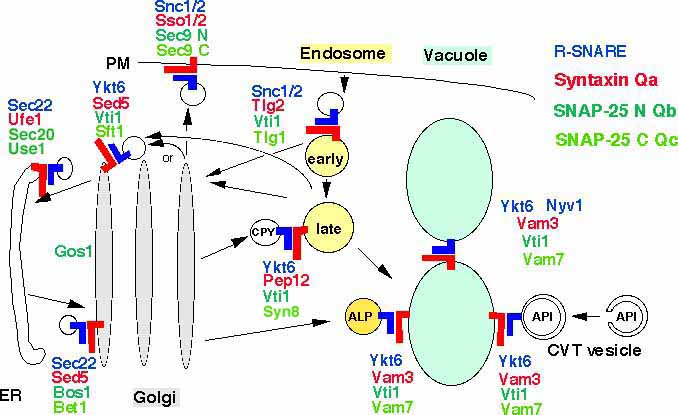
We are interested in the role of SNARE proteins in yeast membrane traffic. We identified a SNARE with low homologies to other SNAREs which functions in retrograde traffic from the Golgi to the ER. It localized to the ER and was therefore named Use1p (unconventional SNARE in the ER, details). The SNARE motifs of Use1p and Sec20p are so divergent that they can't be assigned to either Qb- or Qc-SNAREs.
SNAREs in yeast membrane traffic

The Yeast Q-SNARE Vti1p and the R-SNARE Ykt6p interact with three different syntaxin related Q-SNAREs in several transport pathways
We concentrate on the Qb-SNARE Vti1p and the R-SNARE Ykt6p. SNAREs were thought to determine specificity in vesicular transport. However, Vti1p interacts functionally with three different t-SNAREs in at least four different transport steps. Vti1p functions with the endosomal t-SNAREs Pep12p in traffic from the trans Golgi network to an endosomal compartment and with the cis-Golgi t-SNARE Sed5p in retrograde traffic to the cis-Golgi (details). Vti1p is also required for three different biosynthetic trafficking pathways to the vacuole, the equivalent of the mammalian lysosome and for homotypic vacuolar fusion. Vti1p interacts with the vacuolar t-SNAREs Vam3p and Vam7p in all vacuolar trafficking steps (details). Ykt6p is the R-SNARE in these three SNARE complexes (details). The ability of Vti1p and Ykt6p to mediate multiple fusion steps requires additional proteins to ensure specificity in membrane traffic. The mutations vti1-Q158R and ykt6-R165Q were introduced into the 0 layers to study its role in SNARE complexes. Cells expressing only ykt6Q were not viable. vti1R cells had severe defects in several transport steps. These data indicate that the arginine in the 0 layer of Ykt6p is essential and that an additional arginine in the 0 layer interferes with function (details).
We are currently investigating the role of the N-terminus of Vti1p in yeast membrane traffic. We identified Ent3p as the first protein interacting with this domain (details). Ent3p belongs to the family of ENTH domain proteins which function in the formation of clathrin coated vesicles. Synthetic defects suggest that Vti1p and Ent3p cooperate in transport from the TGN to the prevacuolar endosome.
Whereas a single VTI1 gene is present in the yeast genome two different Vti1p homologs were identified mammals and three in plants. It seems likely that the plant homologs Arabidopsis AtVTI1a and AtVTI1b have different roles in membrane traffic. Upon expression in yeast AtVTI1a can function with Pep12p in Golgi to endosomal transport whereas AtVTI1b can function with Vam3p in traffic to the vacuole but not vice versa (details).
Mammalian SNAREs
Human vti1b can replace yeast Vti1p in certain membrane transport steps if expressed in yeast, indicating that it may have similar functions in mammalian membrane traffic (details). We are currently studying the role of Vti1a and Vti1b in mammalian membrane traffic, their localization and their SNARE partners.
Vti1a and vti1b have a distinct but overlapping localization. Vti1a is found predominantly on the Golgi and the TGN, vti1b mostly on tubules and vesicles in the TGN area and on endosomes (details).
Localization of SNAREs in mammalian cells
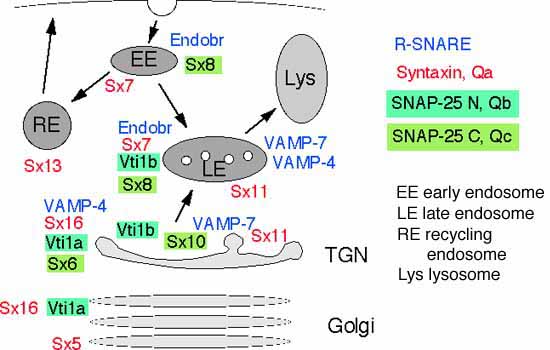
We identified the brain specific splice variant vti1a-b which is localized to synaptic vesicles throughout their life cycle (details). Our data suggest that vti1a-b is not involved in exocytosis but in a separate SNARE complex with VAMP-4, syntaxin 16 and syntaxin 6 (details). This complex probably functions in a membrane fusion step during recycling or biogenesis of synaptic vesicles. It may be required for fusion with early endosomes during recycling because antibodies against vti1a inhibit fusion of early endosomes in an in vitro assay (details). Vti1a has also been implicated in intra Golgi traffic (Xu et al. 1998 J. Biol. Chem. 273, 21783).
Vti1b forms a SNARE complex with syntaxin 7, syntaxin 8 and the R-SNARE endobrevin which shares common structural features with the neuronal SNARE complex. Antibodies against any of the components inhibit fusion of late endosomes in vitro and block lysosomal degradation of EGF in vivo (details).
Common structure of SNARE complexes
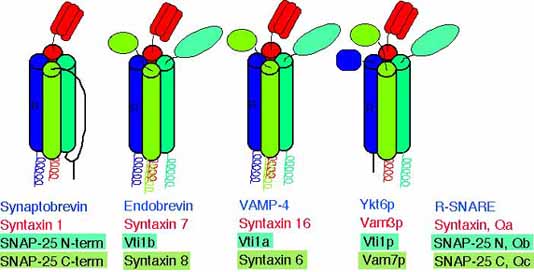
Our data indicate that the SNARE complexes containing the different Vti1 proteins and three additional SNAREs have a similar structure as the complex consisting of synaptobrevin, syntaxin 1 and SNAP-25 despite little similarities in the amino acid sequences.
The N-terminus of vti1b interacts with the ENTH domain of enthoprotin/CLINT/epsinR indicating that this interaction between a SNARE and an ENTH domain is conserved between mammals and yeast(details). Both, epsinR and Ent3p are involved in traffic between the TGN and endosomes. EpsinR and Ent3p may sort vti1 proteins into budding vesicles.
We are generating knock out mice deficient for vti1b to obtain an animal model for analyzes of defects in transport to the lysosome in the whole animal and in specific tissues. Vti1b deficient mice had reduced amounts of syntaxin 8 protein while the amounts of syntaxin 7 and endobrevin did not change. These data indicate that vti1b was specifically required for the stability of a single SNARE partner. Most vti1b deficient mice were indistinguishable from wild type mice. About 20% of the vti1b deficient mice were smaller and accumulated multivesicular bodies and autophagosomes in hepatocytes (details).
last modification Dezember 2004