|
O P A R - Open Architecture Particle in Cell Simulation - Version 3.0
Plasma simulations with dust particles
|
|
O P A R - Open Architecture Particle in Cell Simulation - Version 3.0
Plasma simulations with dust particles
|
| A n-dimensional grid of arbitrary type to hold, densities, fields, boundary values etc. Unlike C-arrays the grid size specifies the maximum index of the elements. This means that the grid contains one element more in each direction than the equivalent C-array. Elements are from (0,0,...,0) to (size[0], size[1], ... , size[n]). The implementation is mostly independent on the dimension of the system because it makes use of the dimensionality dependent Position and GridPosition classes | |
| An abstract base class for parameters. There is only one abstract method: Rebuild. This method has to rebuild the data from the input file and return the next token of th input file in a string | |
| Base class for reading a subclass of the Task class from the input. A Task class is read from the input file by invoking the creating the object and calling its Task::Rebuild method. The parameters of the newly built Task have to be enclosed in curly brackets. The new Task is then attached to its 'parent'. The abstract method New should actually create the object. ParameterTask and DiagnosticTask overwrite this method | |
| An implementation of ParameterTask_BASE for reading in Diagnostics. A Diagnostic Task is special, because it also needs a pointer to the data it is supposed to diagnose. The data is of type OBJECT | |
| An implementation of ParameterTask_BASE for reading in most Tasks. The New method is overwritten to create the Task object. For normal Tasks the default constructor ca be called | |
| Single value paramter of some type. This reads in a single value of some type TYPE from the input file and stores it under a given address. If the parameter is not encountered in the input, a default value is set | |
Solves Poisson equation using a multigrid method for 2D and 3D simulations. The Poisson equation is given by  where where 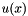 is the unknown scalar field (plasma potential). The outer boundary condition is set to zero, this means on the very outer grid points of the simulation box is the unknown scalar field (plasma potential). The outer boundary condition is set to zero, this means on the very outer grid points of the simulation box  is set. The grid points on the surface of the dust particle(s) is set as a inner boundary condition according to the surface potential of the dust particle(s), calculated from the dust charge, see Potential. This inner boundary condition is time-dependent and can be different in the next timestep, because the charge of the dust particle(s) can change. To follow the spherical surface, reduced grid spacings are used here, which are calculated und stored in the beginning of a simulation, see ClosedGeometry. Periodic boundary conditions are not implemented. For 1D simulations a tridiagonal matrix solver is implemented is set. The grid points on the surface of the dust particle(s) is set as a inner boundary condition according to the surface potential of the dust particle(s), calculated from the dust charge, see Potential. This inner boundary condition is time-dependent and can be different in the next timestep, because the charge of the dust particle(s) can change. To follow the spherical surface, reduced grid spacings are used here, which are calculated und stored in the beginning of a simulation, see ClosedGeometry. Periodic boundary conditions are not implemented. For 1D simulations a tridiagonal matrix solver is implemented | |
| A simple pointer wrapper. Wraps a pointer, so that it can be stored in an STL container. The current implementation has however little advantages | |
| Fundamental class from which all the classes that carry out simulation calculations are derived. The Task contains information of when it is to be executed. Also every object instantiating Task has a name. A Task can also include subtasks which are executed whenever the 'parent' Task is executed | |
| Calculates and stores the charge density on the grid points in the simulation box. The calculation is done by requesting from all the Species to calculate their particle density and then adding up the results | |
| This is the base class for any diagnostic task inside another Task. It doesn't do much but allows the user to specify a DiagPort object by it's name in the setup file | |
| Template Diagnostic to write out a grid of any type. The TYPE has to be cast-able to double | |
| Diagnostics for just one line of a 2d- or 3d-grid | |
| Template Diagnostic to write out any single value type. The TYPE has to define the operator /(T,double) the result of which has a matching DiagPort::Out method | |
| Diagnostic to write out the trajectory of a single particle of a Species | |
| Abstract class for directing diagnostics to some output. The DiagPort defines an abstract interface for directing diagnostic data to some output. At the moment only the File class implements this interface | |
| Implements a DiagPort for writing data to a file. The Series parameter of the DiagPort has the following meaning: if set to true (1) then only one file is produced with all new data appended at the end. If set to false (0) the file name given by the file parameter is parsed, replacing the first occurence of "#t" with the current time step. Then for each diagnostic output a new file is created. If the file name doesn't contain "#t" then everytime an output is created the old data is overwritten | |
| Task that implements an electrostatic E-field. It consists of a grid with n-dimensional entries. The field is calculated by taking the derivative of the electrostatic potential. The electric field in the environment of the dust particle(s) is calculated with the same method as in the poisson-solver using reduced grid spacings to follow the spherical dust particle surface | |
| An abstract Task class for the initial generation of particles | |
| A class derived from Generation to homogeneously Generate particles. The particles have directed thermal velocity components according to Maxwells velocity distribution | |
| Defines an abstract interface for geometry definitions. These may include open geometries, closed geometries or complex box shapes | |
| Implements a Geometry representing a closed simulation box | |
| Accelerates and moves particles. Simplest non relativistic, electrostatic case. It is also possible to implement a constant and homogeneous magnetic field Bz in z-direction. This simple movement class uses the simple integration of the non relativistic Newtonian equations of motion with only an electric field. After that the influcence of the constant and homogeneous magnetic field will be considered geometrically, because the magnetic field do not cause energy change to the particle, but spiral movement along the magnetic field lines in z-direction. This means: First of all a new position of the particle will be calculated according to the Newtonian equation with only an E-field. Then this movement between the old and the new position is considered with a constant and homogeneous B-field in z-direction to find the position that is set in the next simulation step. The constant magnetic flux density Bz is determinated via the most probably gyration radius of electrons rg which is set in the ElStaticNorm. If no gyration radius is set there, no magnetic field is set here, Bz=0. Bz can only be considered in 2D and 3D simulations | |
Accelerates and moves particles for 1D simulations. Extends the simplest non relativistic, electrostatic case to spherical geometry. This simple movement class uses the simple integration of the non relativistic Newtonian equations of motion with only an electric field. The motion is implemented in spherical geometry. Only 1D spherical geometry is implemented. The second velocity component is interpreted as 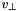 | |
| Abstract interface for normalization classes. These classes allow the user to provide normalized values in the input file. These will then be transformed by the normalization class into the physical non-normalized quantities | |
| Normalization used for electrodynamic simulations. Please note: This class can not be used together with dust particles, because no update has been done. E-Dynamic consideration was not yet planed. This class had to be completely reworked before using | |
| Normalization used for electrostatic simulations | |
| A base class for potential classes | |
| Class for the potential in simulations with dust particle(s) | |
| A specialized Task which organizes all the other Tasks in a Process and allows reading the Tasks from a setup file. This class is designed for running as the parent of all other classes in a process. It keeps track of the current time step in nCycle and has a special method Run that Runs the process | |
| Simulation specializes Process and is supposed to implement the the master process. The Simulation class takes care of registering all the Tasks that can be read from the input file at top level. It also provides some routines for Drawing and Interactivity | |
| Base class for Species. The charge density has to access methods MakeRho and Rho. To allow different implementations of the Species class, we create this interface class | |
| This class implements an individual particle species. A Species class contains all the positions and velocities of a particle species. It also contains direct links to a Wall Object and a grid containing the particle density. Normally a species will contain at least a Movement sub-Task and either a Generation sub-Task or some Task that will create particles during run time | |
| A simple totally absorbing wall in all boundary directions, absorbing dust particle(s) and insertion of particles at all boundary grid cells. Absorbing wall and dust particle: Test all the particles of all the Species if they are outside the simulation box or inside the dust particle. If so the particle is removed from the simulation. If the particle is absorbed by the dust particle, the charge (-+1) is added at the dust particle(s) charge(s). Insertion of particles at all boundaries: Number of inserted particles per boundary cell is calculated from the current density per grid cell: j = np*Vm/dx, where Vm is the average thermal velocity of the particles according to Maxwells velocity distribution | |
| Velocity_ holds the template of a 3-dimensional 'vector' |
 1.8.1.1
1.8.1.1